Researchers from the Center for Nanoscience at the University of Jyväskylä in Finland, developed a calculation model It could facilitate the use of nanomaterials in biomedical applications. Their machine learning framework is able to predict how proteins interact with ligand-stabilized gold nanoclusters, a material widely used in bioimaging, biosensing, and targeted drug delivery.
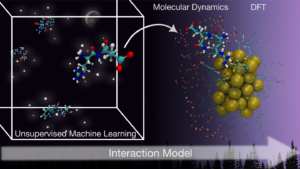
By integrating machine learning and atomistic simulations, the researchers developed a new interaction model that reveals the chemical laws that guide protein binding to gold nanoclusters. Image: Brenda Ferrari.
Gold nanoclusters are used for bioimaging by taking advantage of their natural fluorescence. For example, the material could be coated with molecules that are attracted to cancer cells, allowing doctors to pinpoint the location of tumors. Gold nanoclusters that are engineered to change color when in contact with proteins are also used to detect biomarkers. Unlike most nanoparticles, gold nanoclusters are small enough to pass through the kidneys and be excreted from the body in the urine, making them safer than other nanoparticles.
Molecular dynamics simulations are a common approach to studying protein-nanoparticle interactions, but computational modeling can be challenging due to the complexity of the interactions between each particle and between the particles and the environment.
“MD simulations of all these structures are feasible but an expensive venture. On the other hand, for peptides with more than four amino acids, this is not a feasible approach. The application of machine learning techniques appears to be a promising approach to address this problem,” the researchers wrote in their paper. Published in a magazine totalling.
Existing studies predicting how proteins will interact with nanoparticles often focus on isolated cases, and researchers lack a unified predictive model to guide their designs. To address this, researchers at the Nanoscience Center developed a clustering-based machine learning framework that identifies the chemical principles involved in the adsorption of biomolecules onto gold nanoclusters.
“This model determines which amino acids have higher or lower priority for binding to gold nanoclusters and identifies the specific chemical groups involved in these interactions,” said Brenda Ferrari, a postdoctoral researcher at the university. stated in the press release.
This framework can be extended beyond peptides and may provide broader insights into the interactions of protein and gold nanoclusters, accelerating the screening process. Using this model, the design of protein-gold nanocluster compounds could become more efficient by allowing for faster screening of specific functions and properties of proteins.
“Our goal was to build a model that was generalizable, rather than just explaining one specific system,” Ferrari says. Although we will continue to push the limits, we already have a model that broadly describes the interaction of protein and gold nanoclusters and can be extended to support the development of smarter nanomaterials for biomedical applications, she continues.




