To comprehensively evaluate our method, we utilized generated barcodes to perform image searching across four distinct datasets, encompassing both medical and non-medical domains. Furthermore, we diversified the selection of conventional network types for the purpose of feature extraction. It aims to demonstrate the model’s flexibility in handling various data types and its potential for use in practical situations. The whole process is to extract the features from the experimental datasets and then use them. This section describes the details of the experiments that were conducted. Importantly, we adhere strictly to standard machine learning practices by performing optimization solely on the training sets of the datasets. The test sets were used exclusively to evaluate the final performance of the optimized barcodes to ensure that no information from the test sets influenced the optimization process. This distinction underlines the validity of our approach and its ability to generalize to unseen data. This section describes the details of the experiments that were conducted.
Datasets
TCGA
TCGA dataset offers public digital pathology repositories71, which are first initialized by the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI). The TCGA database contains 30,072 histopathological whole slide images (WSIs) of 32 cancer subtypes and corresponding metadata. This pathology dataset has been extensively used in deep learning for cancer research. In addition, the annotations are at the WSI level (no pixel-level delineations) along with morphology, primary diagnosis, and tissue or organ of origin metadata72. Due to the gigapixel size of WSIs (often much larger than 50,000 by 50,000 pixels), analyzing and processing steps could be a challenge for data scientists. One way to overcome this is to extract WSI portions, called patches, of ordinary dimensions, say 1,000 by 1,000 pixels, with less computational complexity. Since we needed to apply our model to extracted features, we collected the features of TCGA images extracted by KimiaNet, a pre-trained DenseNet-121 by Riasatian et al.73 to simplify feature extraction steps. In this paper, tissue patches derived from WSIs with \(1000\times 1000\) pixels were fed to KimiaNet to extract deep features. However, the average feature pooling introduced in74 can lead to one feature vector of length 1024 for the entire WSI.
Due to the curse of dimensionality in feature space size of 1024, several works such as Rasool et al.75 recommended doing feature selection using GA, which significantly improves accuracy due to redundancy reduction. Recent work by Asilian et al.76 proposed an evolutionary multi-objective optimization framework to generate highly compact and discriminative WSI representations. Their method begins by extracting patch-level features using KimiaNet and applies a two-stage feature selection strategy. To further explore the impact of barcoding and CBIR on reduced feature space, we also experiment with our proposed method on the reduced feature subset derived by Asilian et al.76 in addition to KimiaNet extracted full features.
In our experiments, we utilized the extracted deep features for multiple cancer subtypes in the TCGA dataset. Details of the dataset split used in training, validation, and testing are summarized in Table 1.
COVID-19 radiography
In the second test case, we employed a publicly available COVID-19 dataset77,78, including four classes: COVID-19, Pneumonia, Lung Opacity, and Normal. We then trained the ResNet-50 architecture from scratch for feature extraction. ResNet-50, recognized for its deep network layers, has residual blocks that allow for the training of large networks while overcoming gradient-related issues. These blocks, when combined with convolutional and pooling layers, capture increasingly complex visual characteristics. The architecture culminates in global average pooling and fully connected layers, which turn these features into a compact categorization representation. Finally, the obtained feature vectors are of length 128 as the input of the image search system79,80.
Fashion-MNIST
The third dataset, Fashion-MNIST, is a popular non-medical machine learning and computer vision benchmark. It consists of grayscale photographs of T-shirts, trousers, pullovers, dresses, coats, sandals, shirts, shoes, purses, and ankle boots from ten various fashion categories in a 28\(\times\)28 pixel representation81,82. In this study, we fine-tuned EfficientNet-B083 by adding a customized feature extractor network, including global average pooling to summarize spatial information, followed by a dropout layer to prevent overfitting. Lastly, a dense layer with a softmax activation function to align with the Fashion-MNIST dataset classes. Subsequently, we utilized the 1280-length feature vector obtained from the second-to-last layer as non-medical test cases for our CBIR system.
CIFAR-10/100
The CIFAR-10 and CIFAR-100 datasets consist of 32\(\times\)32 color images in 10 and 100 distinct classes84. In order to obtain the requisite feature vectors, we conducted fine-tuning on DenseNet-12185, augmenting it with a global average pooling layer and flattening layers, accompanied by a sequence of Dense Layers configured with output sizes of 1000 and 10. Similar to previous datasets, we employed the features extracted from the Dense layer preceding the final layer for our specific objective.
Implementation details
We compare our proposed CGA method to various hashing approaches in image retrieval, including dHash25 and DFT-Hash are as baselines for order-sensitive hashing methods, and other aHash30, MinMax22, LBP86and, ITQ21are non-learnable hashing methods. Learnable hashing methods have recently been widely used due to the rise of neural networks, such as, DHN26, DPSH87, Quantization29, LSH32, CSQ28, DTSH27, and DSH23. To create a fair comparison, we use pre-trained DNNs such as KimiaNet and DenseNet-121 for the TCGA dataset, ResNet-18 for the CIFAR-10 and CIFAR-100 datasets, EfficientNet-B0 for the Fashion-MNIST dataset, and ResNet-50 for the COVID-19 dataset as backbone models and fine-tune on the classification task using the cross entropy loss function. Then, we use their convolutional layer to extract feature representation by removing the classification head. Therefore, all hashing methods use the same inputs as extracted features and transform them into binary values. We then evaluate the methods on three metrics using training data as a database and test data as a query after transforming to barcodes. The test set is only used in the evaluation phase to query the test barcodes on the whole training barcode database and return the most related samples in the database. For learnable neural network-based hashing methods, the number of epochs is set to 100, and the batch size is set to 512. In our CGA method, the number of iterations is set to 100 and population size \(NP=100\) to keep the consistency in maximum number of function calls as \(Max_{FE} = 10,000\). Furthermore, the crossover probability rate is set to 0.9, and the mutation probability rate in inversion mutation is set to 0.9. Due to the inherent stochasticity of all hashing methods including ours, each method is independently repeated 15 times with different random seeds. To evaluate statistical significance, we perform paired t-tests between each baseline method and our proposed CGA approach. The resulting p-values are reported in parentheses (e.g., \((p\text {-value})\)) alongside each metric in the tables.
Results and analysis
In this section, we discuss the experiment carried out to evaluate the proposed barcode optimization algorithm. The experiments include a comparison of the hashing techniques in the generation of barcodes for image search tasks, which is assessed across disparate types of datasets to show how our proposed method affects the data retrieval performance against other methods on F1-score, Precision@\(k=10\), and mAP evaluation metrics in the following analyses.
Retrieval performance on TCGA
We evaluated the effectiveness of the proposed CGA in optimizing the ordering of features extracted from two pre-trained deep neural networks, KimiaNet and DenseNet-121, using the TCGA histopathology dataset. CGA functions as an evolutionary strategy for discovering the most informative arrangement of features, improving the quality of binary representations used for image retrieval and classification. Its performance is comparable to traditional binary encoding schemes, unsupervised hashing techniques, and deep supervised hashing models. The evaluation considers F1-score, Precision at top-\(k=10\) retrieval (Precision@k), and mean average precision (mAP), across both complete and reduced feature sets. In the following paragraphs, each TCGA sub-dataset’s barcoding retrieval performance are analyzed individually based on feature extraction types.
Retrieval Performance on TCGA Extracted Features by KimiaNet In the first experimental setting, we evaluate all methods on the KimiaNet-Pulmonary dataset using full feature vectors. As shown in Table 2, our proposed CGA-dHashmethod outperforms all baselines by achieving the highest Precision@k (0.8743 ± 0.01) and mAP (0.8620 ± 0.00), while also ranking second in F1-score (0.8861 ± 0.01), just behind DFT-Hash (0.8954 ± 0.00). CGA-dHash attains the highest overall average score (0.8741) and the best average rank (1.3), indicating a consistent advantage across all three metrics. CGA-DFT also demonstrates robust performance, placing among the top methods with strong scores across the board F1-score of 0.8722 ± 0.01, Precision@k of 0.8522 ± 0.01, and mAP of 0.7963 ± 0.01–yielding an overall average of 0.8402 and an average rank of 4.3. These results confirm that our CGA-based optimization is highly effective in refining feature permutations to enhance semantic consistency within neighborhood retrievals. This leads to notable improvements in both image retrieval precision and classification quality. Additional results on the rest of TCGA sub-datasets are presented in Supplementary Tables 8 to 14.
Retrieval Performance on TCGA Extracted Features by KimiaNet with Fine Selection To assess CGA’s robustness when working with a smaller subset of features selected by Asilian et al.76, we apply feature selection to reduce the dimensionality of the KimiaNet’s Endocrine representations. As presented in Table 3, CGA-dHash consistently outperforms all baseline methods, achieving the best results in F1-score (0.9935 ± 0.01), Precision@k (0.9898 ± 0.01), and mAP (0.9798 ± 0.01). It also achieves the highest average score (0.9877) and the top average rank (1.0), demonstrating its strong and stable performance under feature-restricted settings. In contrast, CGA-DFT shows moderate results with F1-score of 0.8776 ± 0.04, Precision@k of 0.8628 ± 0.04, and mAP of 0.7969 ± 0.05, resulting in an average score of 0.8458 and an average rank of 12.7. Notably, while some deep supervised hashing methods, such as ITQ21 and DFT-Hash, also maintain relatively strong scores, several others, including contrastive or quantization-based objectives, deteriorate significantly in this setting. This contrast highlights the ability of CGA to remain effective and adaptable regardless of the number of features, whereas many neural-based hashing methods are more sensitive to dimensionality reduction. Additional results on the rest of TCGA sub-datasets are presented in Supplementary Tables 1 to 7.
Retrieval Performance on TCGA Extracted Features by DenseNet-121 The analysis on DenseNet-121 extracted features presented in Table 4 provides compelling evidence for the effectiveness of the proposed CGA methods on various deep neural networks. Specifically, CGA-dHash achieves the best results across all three evaluation metrics: F1-score (0.7070 ± 0.03), Precision@k (0.6536 ± 0.01), and mAP (0.5719 ± 0.01). This leads to the highest average score (0.6441) and the lowest average rank (1.3), indicating robust and consistent performance across different retrieval quality indicators. In contrast, CGA-DFT performs moderately with an average of 0.5706 and an average rank of 7.0, showing lower adaptability to DenseNet-121 derived features compared to CGA-dHash. Interestingly, most traditional and deep hashing methods, including Quantization29, DSH23, and pairwise-based approaches–exhibit considerably weaker performance in this setting. These findings suggest that DenseNet-121 features pose a greater challenge to conventional hashing strategies. However, CGA-dHash demonstrates superior adaptability, effectively identifying more informative feature permutations for retrieval and classification. Additional results on the remaining TCGA sub-datasets are presented in Supplementary Tables 15 to 21.
Across all experiments, CGA delivers superior performance compared to existing approaches. Unlike deep hashing methods that do not directly align their learning objectives with binary encoding or retrieval metrics, CGA explicitly optimizes the arrangement of features to maximize similarity-based rankings. This leads to improved structure in the binary space and better alignment with downstream evaluation objectives. Moreover, CGA operates independently of the network architecture and is equally effective on both full and reduced feature representations. These characteristics make it a practical and efficient solution for enhancing compact binary representations, particularly in retrieval tasks where interpretability, memory efficiency, and precision are essential.
Retrieval performance on CIFAR-10/100
To evaluate the generalizability of the proposed CGA, we extend our experiments to two widely used natural image benchmarks: CIFAR-10 and CIFAR-100. These datasets include object classes with diverse visual characteristics and serve as a standard for image retrieval and classification performance. Each method is evaluated over 15 independent runs, and results are reported using three standard metrics: F1-score, Precision at top-k retrieval (Precision@k), and mean average precision (mAP).
Retrieval Performance on CIFAR-10 The results in Table 5 indicate that CGA demonstrates strong performance across all metrics, particularly in terms of F1-score (0.9337) and mAP (0.9274), which positions it competitively among the leading methods. The aHash30method achieves the top performance for all three individual metrics (F1-score: 0.9362, Precision@k: 0.9353, and mAP: 0.9489), consequently obtaining the lowest average rank (1.0). However, CGA outperforms many other traditional hashing and optimization methods, such as DPSH87, DSH23, and DHN26, which shows CGA’s capacity to maintain robustness and stability in performance. Despite not surpassing aHash30’s peak scores, CGA’s consistently high performance emphasizes its potential as a reliable and effective method for CIFAR-10 classification tasks.
Retrieval Performance on CIFAR-100 The analysis based on the more challenging CIFAR-100 dataset (Table 6) reveals distinct insights into the methods’ capability to manage higher complexity due to increased classes and finer-grained categorization. CGA attains the highest F1-score (0.7496) compared to all other methods, clearly indicating its superior ability to distinguish among a higher number of classes. In terms of overall average performance (Avg = 0.6582), CGA maintains competitive performance but slightly trails the leading methods LBP86(0.7235 Avg) and aHash30(0.7222 Avg). While LBP86dominates in Precision@k (0.7359) and mAP (0.6843), CGA still demonstrates commendable robustness, substantially outperforming other hashing approaches (e.g., DPSH87, DSH23, DHN26, and Quantization29), which significantly degrade in performance. Therefore, CGA showcases considerable potential and adaptability in handling datasets characterized by higher complexity and class diversity.
Cross CIFAR-10/100 Datasets Observations Across both natural image benchmarks, CGA demonstrates competitive performance and consistently outperforms a number of conventional and supervised hashing models. Its ability to discover meaningful feature orderings that preserve local similarity and neighborhood structure makes it a valuable tool in binary encoding tasks. Unlike neural-based hashing models, which often fail to generalize when trained on limited or complex distributions, CGA directly targets evaluation metrics and remains adaptable to various feature distributions and backbone architectures. These results confirm that the ordering of features plays a critical role in the performance of binary representations and that optimization via combinatorial search can lead to measurable improvements in both retrieval and classification tasks.
Retrieval performance on Fashion-MNIST
To further assess the adaptability of CGA across different visual domains, we evaluate its performance on the Fashion-MNIST dataset. This dataset, although grayscale and lower in resolution, provides a challenging classification and retrieval task due to subtle inter-class differences in clothing categories. As in previous experiments, we report F1-score, Precision at top-k (Precision@k), and mean average precision (mAP), averaging over 15 independent runs.
As presented in Table 7, CGA achieves the best overall performance across all metrics. Specifically, it obtains the highest F1-score (0.8623 ± 0.01), Precision@k (0.8078 ± 0.00), and mAP (0.6217 ± 0.00). These values resulted in the top average score (0.7639) and the lowest average rank (1.3), confirming the method’s superiority in balancing classification accuracy and retrieval quality. In comparison, the aHash30 encoding approach achieves an F1-score of 0.8589 ± 0.07 and Precision@k of 0.7997 ± 0.00, ranking second overall. While LBP86and ITQ21methods perform competitively in terms of Precision@k, they fall short in mAP, indicating lower retrieval consistency across the feature space. Supervised deep hashing methods consistently underperform, with models such as Quantization29and DSH23producing extremely low scores across all metrics. This is especially notable in mAP, where the best-performing deep hashing variant DPSH87achieves only 0.2765 ± 0.00. These results further emphasize the limitations of learning-based hashing methods when applied to datasets with low inter-class variability and limited input modalities. The strong performance of CGA on Fashion-MNIST demonstrates its flexibility across both natural and synthetic visual domains. Unlike neural-based binary encoding approaches, which often fail to generalize in low-complexity settings, CGA directly optimizes for evaluation-driven objectives by discovering informative permutations in feature space. Its ability to outperform supervised methods without requiring training or labeled pairs underscores its effectiveness as a lightweight and data-efficient encoding solution. These findings further support the conclusion that feature ordering plays a vital role in retrieval-based performance and that combinatorial optimization strategies such as CGA provide a robust alternative to deep binary representation learning.
Retrieval performance on COVID-19
To investigate the effectiveness of CGA on real-world medical data with challenging feature distributions, we conduct experiments on the COVID-19 dataset. This dataset introduces additional complexity due to data imbalance and heterogeneity in the imaging conditions. As shown in Table 8, the aHash30 encoding approach yields the best average performance, achieving the highest ranking among all methods. It is followed closely by LBP86and ITQ21, which also showed strong results in both retrieval and classification tasks. dHash25 performs surprisingly well given its minimalistic design, reflecting the potential advantage of feature magnitude trends even in high-noise clinical datasets. CGA produces a competitive average score of 0.7628, placing it within the top-performing group and achieving an average rank of 4.3. While it did not outperform the aHash30 method in this setting, CGA maintains robust and consistent performance across all runs. Compared to many deep supervised hashing models, including Quantization29, CSQ28, and DSH23, which showed severe performance degradation, CGA offers a much more stable and reliable solution. The ability of CGA to remain effective under the conditions of a noisy and imbalanced dataset underscores its potential in clinical applications. Rather than relying on supervised signal propagation or class-specific anchors, CGA leverages the intrinsic structure of the feature space through permutation optimization. This allows it to preserve neighborhood relations without requiring class labels or pairwise training.
Overall, the results on the COVID-19 dataset support the broader observation that feature ordering has a non-trivial influence on binary representation quality. CGA remains a promising and model-independent approach for optimizing such encoding methods across both synthetic and clinical imaging domains.
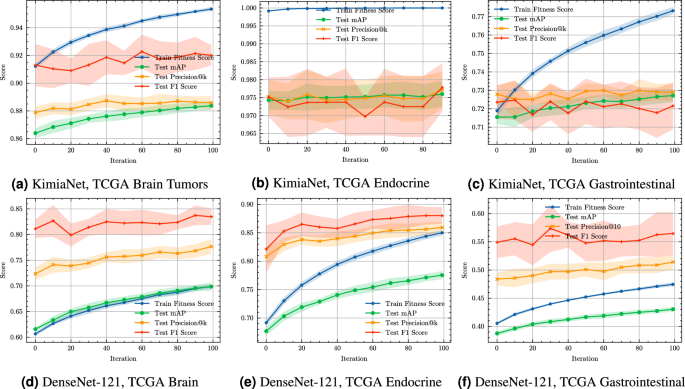
Convergence plots for CGA-dHash barcoding optimization on TCGA dataset features extracted by KimiaNet and DenseNet-121.
Convergence analysis of CGA
To further investigate the optimization dynamics of the proposed CGA method on dHash25, we provide convergence plots on six TCGA sub-datasets derived from KimiaNet and DenseNet-121 feature representations, as shown in Figure 3. These plots illustrate the evolution of the training fitness score (used as the objective in the evolutionary search) alongside three evaluation metrics on the test set: mAP, Precision@k, and F1-score. Convergence plots for additional TCGA sub-datasets including KimiaNet and DenseNet-121 extracted, selected features, and other datasets are shown in
Figures 1 to 4, respectively.
In subplot Figure 3a, corresponding to KimiaNet features on TCGA brain tumor samples, CGA-dHash demonstrates a clear upward trend across all test metrics. Both Precision@k and F1-score steadily improve throughout the iterations, reflecting the model’s increasing ability to preserve neighborhood relationships in the binary space. The training fitness score also rises consistently, indicating successful convergence of the evolutionary search process. The alignment between training fitness and test metrics highlights that the learned permutations are effectively generalizing to unseen data. In contrast, Figure 3b for the Endocrine subset shows relatively flat convergence curves for all test metrics, despite a consistently high training fitness score. This indicates a potential saturation effect or a low variance in the Endocrine representations, where the CGA-dHash is already near-optimal at initialization. Nonetheless, the performance remains stable, suggesting robustness in evolutionary search without overfitting. For the Gastrointestinal data in Figure 3c, CGA-dHash shows steady improvement in the training fitness score, with corresponding but modest gains in the test metrics. Precision@k and F1-score exhibit mild but consistent upward trends, confirming that even in lower-performing settings, the optimization process gradually improves representation quality.
Figure 3d, Figure 3e, and Figure 3f present convergence on DenseNet-121 extracted features from the same tissue types. In Figure 3d, the Brain tumor subset again reflects strong convergence behavior with significant improvements in all test metrics, especially F1-score, which stabilizes above 0.8 after 60 iterations. Figure 3b for the endocrine data shows near-linear improvement in all test scores, demonstrating that CGA-dHash efficiently identifies better permutations over time. Finally, in Figure 3c, while test metrics for Gastrointestinal samples exhibit modest increases, the training curve remains consistently upward, mirroring the behavior observed in KimiaNet extraction.
Overall, these convergence plots demonstrate that CGA-dHash consistently improves retrieval performance over iterations. The strongest convergence is observed in high-variance datasets such as Brain and Endocrine tissues, while in more homogeneous datasets like Endocrine extracted by KimiaNet, CGA-dHash maintains stable performance. These findings provide further evidence that CGA-dHash’s optimization process is both effective and resilient across medical image domains and feature extraction backbones.
Parameter sensitivity analysis
To investigate how different hyperparameter settings influence the performance of our CGA-dHash, we evaluate combinations of the crossover probability rate (CR) and mutation probability rate (MR) across three datasets: DenseNet-121-Brain, DenseNet-121-Endocrine, and DenseNet-121-Gastrointestinal. The results, shown in Table 9, include five key performance metrics: F1 Score, Precision@k, mAP, average score, and average rank. The best results for each metric are highlighted in bold, with the second-best results underlined.
Impact of Crossover Rate (CR) A consistent pattern emerges across all datasets: increasing the crossover rate from 0.1 to 0.9 leads to improved performance across most metrics. In particular, the configuration with \(CR = 0.9\) significantly outperforms others. For example, in the Brain dataset, \(CR = 0.9\) with \(MR = 0.1\) achieves the highest average score (0.7580) and the best average rank (1.0). A similar trend is observed in the Endocrine dataset, where the same setting yields a strong performance (Avg = 0.8383, Rank = 2.0), closely following the top-ranked \(CR = 0.9\), \(MR = 0.9\) configuration. This indicates that a higher crossover rate enhances the algorithm’s ability to explore and recombine promising solutions effectively.
Impact of Mutation Rate (MR) The effect of mutation rate appears to be more dataset-dependent. In the Brain dataset, a lower mutation rate (\(MR = 0.1\)) is more effective when paired with high crossover. However, for the Gastrointestinal dataset, where performance margins are smaller and data is likely noisier, the highest mutation rate (\(MR = 0.9\)) yields the best overall performance (Avg = 0.4998, Rank = 1.3). This suggests that mutation contributes significantly to maintaining diversity and avoiding premature convergence in more complex or less structured datasets.
Cross-Dataset Observations The configuration \(CR = 0.9\), \(MR = 0.1\) consistently performs among the top two across all datasets, establishing it as a strong general-purpose setting. However, \(CR = 0.9\), \(MR = 0.9\) achieves the best average rank on the Endocrine and Gastrointestinal datasets, highlighting the potential benefit of higher mutation rates in certain domains. Notably, settings with low crossover (\(CR = 0.1\)) result in the poorest performance in all cases, reaffirming the importance of effective recombination in CGA.
Based on the empirical evidence, we recommend using a high crossover rate (\(CR = 0.9\)) and mutation rate (\(MR = 0.9\)) across all datasets to increase the convergence.
Cross-dataset performance summary
The results summarized in Table 10 highlight the superior performance of our proposed method, CGA, including its variants CGA-dHash and CGA-DFT, across a diverse set of 28 datasets. CGA, which can be applied to both dHash25 and DFT-Hash feature extractors, achieves the highest average score, computed as a composite of F1, Precision@k, and mAP metrics, in 14 datasets, representing 50% of the evaluated cases. This includes datasets such as KimiaNet-Brain (0.8991), KimiaNet-Endocrine (0.9796), DenseNet-121-Liver (0.7917), and Fashion-MNIST (0.7639), among others. Notably, CGA-DFT, a variant of CGA applied to DFT-Hash, secures the top position in 3 datasets, including KimiaNet-Gastrointestinal (0.7277) and KimiaNet-Melanocytic (0.9365). In total, our proposed method outperforms all competing methods in 14 instances, demonstrating its effectiveness and robustness.
Furthermore, CGA-dHash or CGA-DFT ranks among the top two methods in 22 out of 28 datasets (approximately 79%), reinforcing its consistent high performance. In datasets that do not claim the top spot, it frequently secures the second position, as seen in KimiaNet-Brain with selected features (0.8488) and DenseNet-121-Gynecologic (0.6705). Remarkably, in two datasets, KimiaNet-Gastrointestinal and KimiaNet-Melanocytic, our methods dominate by occupying both the first and second ranks, with average scores differing by less than 0.0015, underscoring the strength and synergy of CGA-dHash and CGA-DFT.
Compared to other methods such as Quantization29, aHash30, and DPSH87, which occasionally achieve top ranks, CGA-dHash surpasses them in frequency and consistency. For instance, aHash30is the best method in 5 datasets, and Quantization is the best method in 3, yet neither matches the 14 top rankings of CGA-dHash and CGA-DFT combined. This dominance is particularly evident across medical imaging datasets (e.g., KimiaNet and DenseNet-121 series) and extends to general datasets like Fashion-MNIST, illustrating the versatility of our approach across different domains and feature representations.
In conclusion, the proposed CGA method, including its variants, definitively surpasses competing methods by securing the highest performance in half of the evaluated datasets and maintaining a top ranking in the vast majority. These results establish CGA as a leading technique for the tasks assessed, validated by its robust and adaptable performance.




